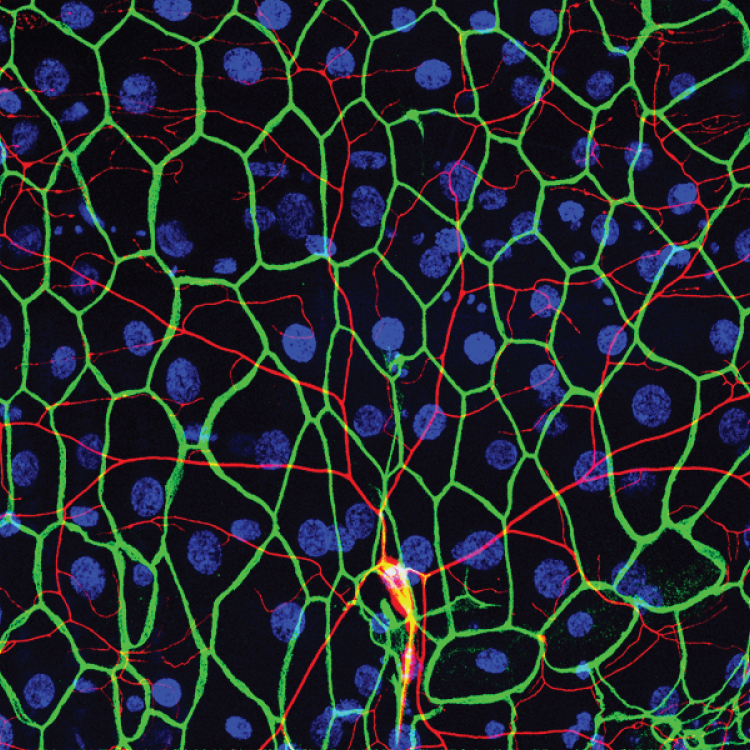
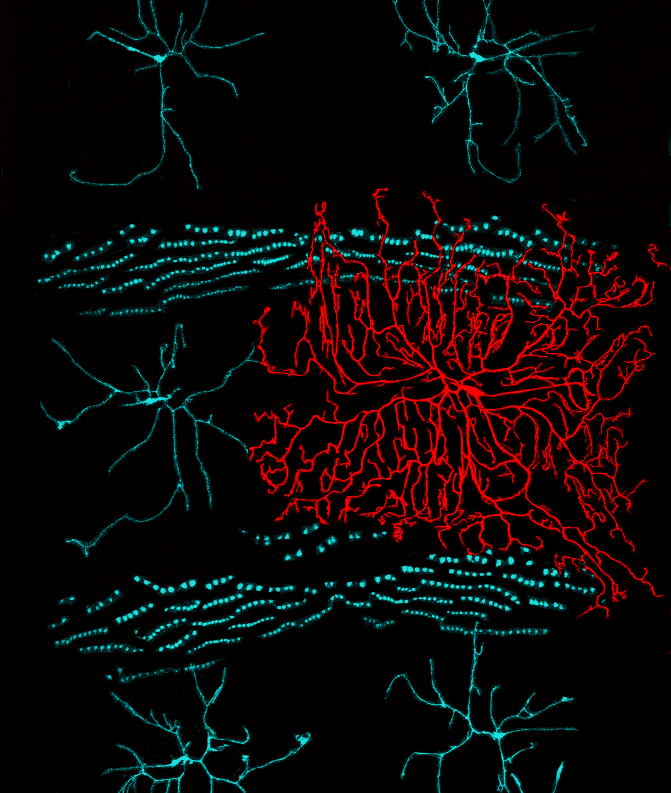
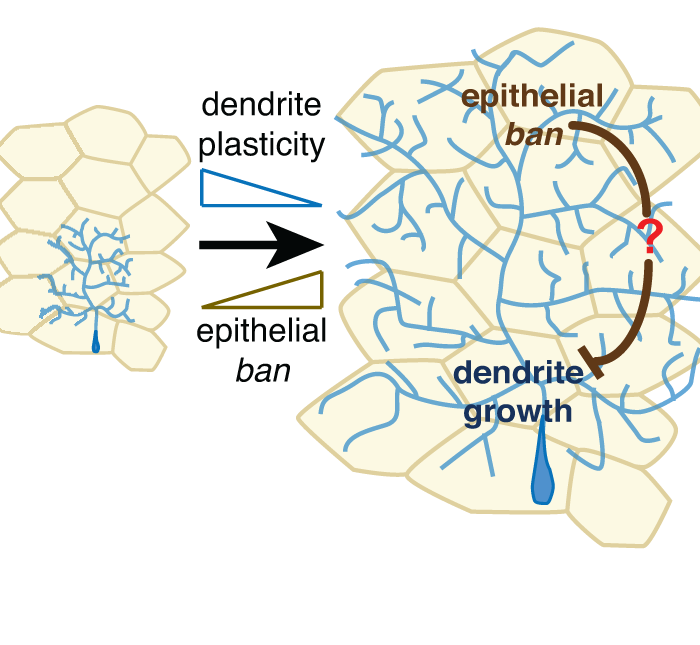
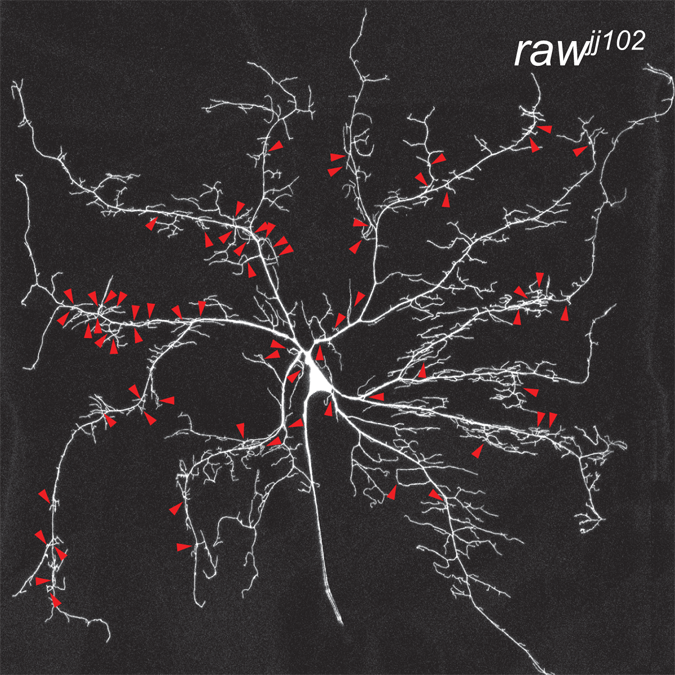
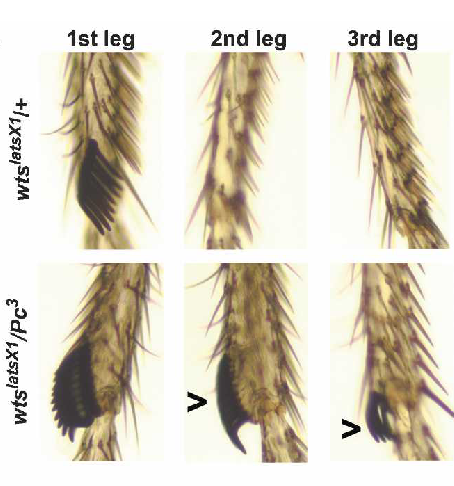
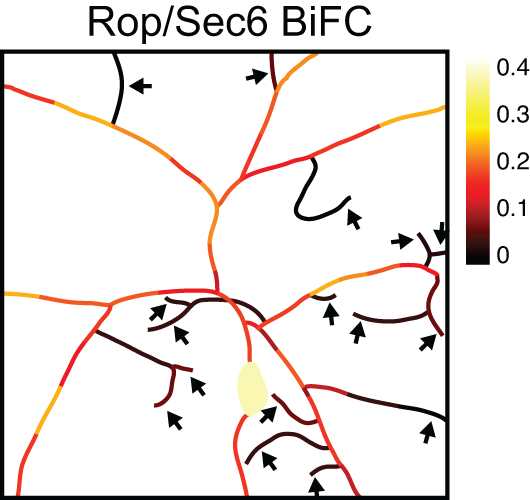
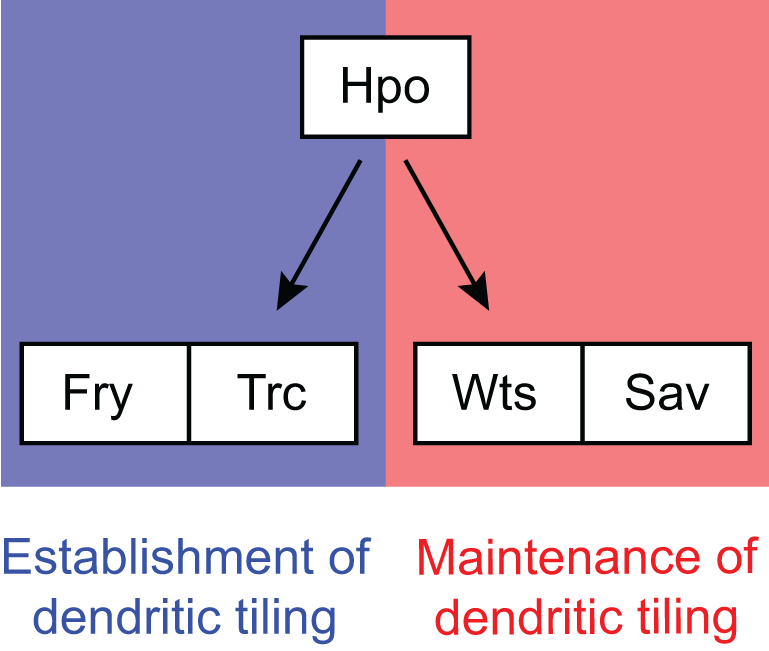
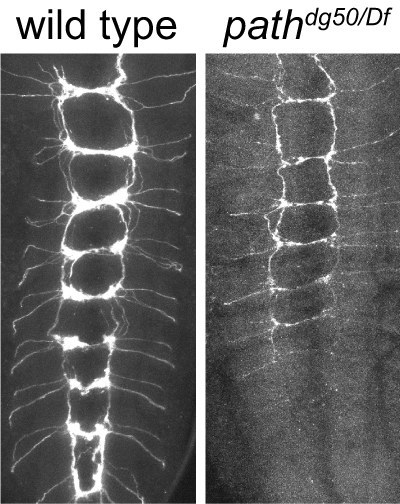
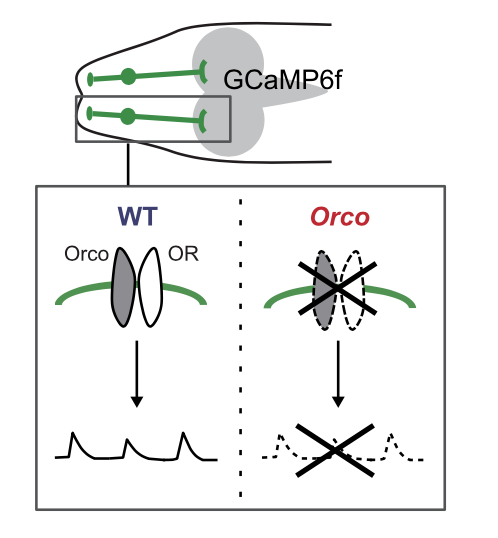
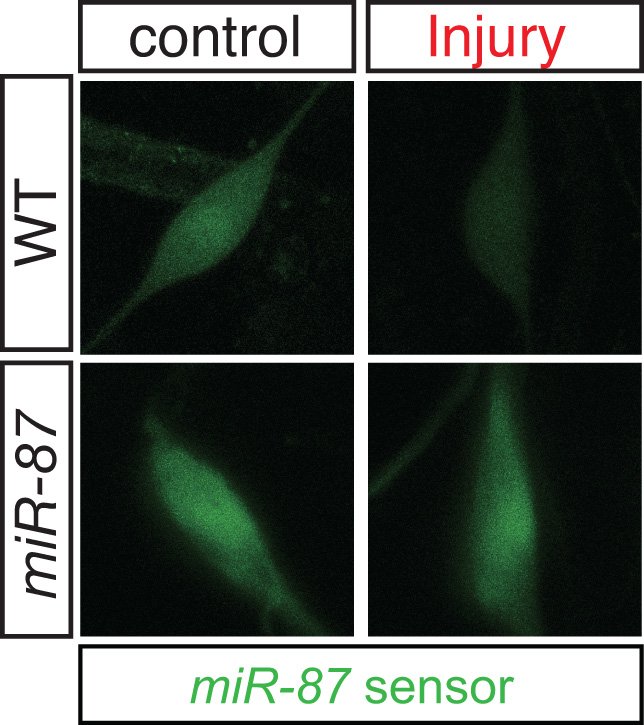
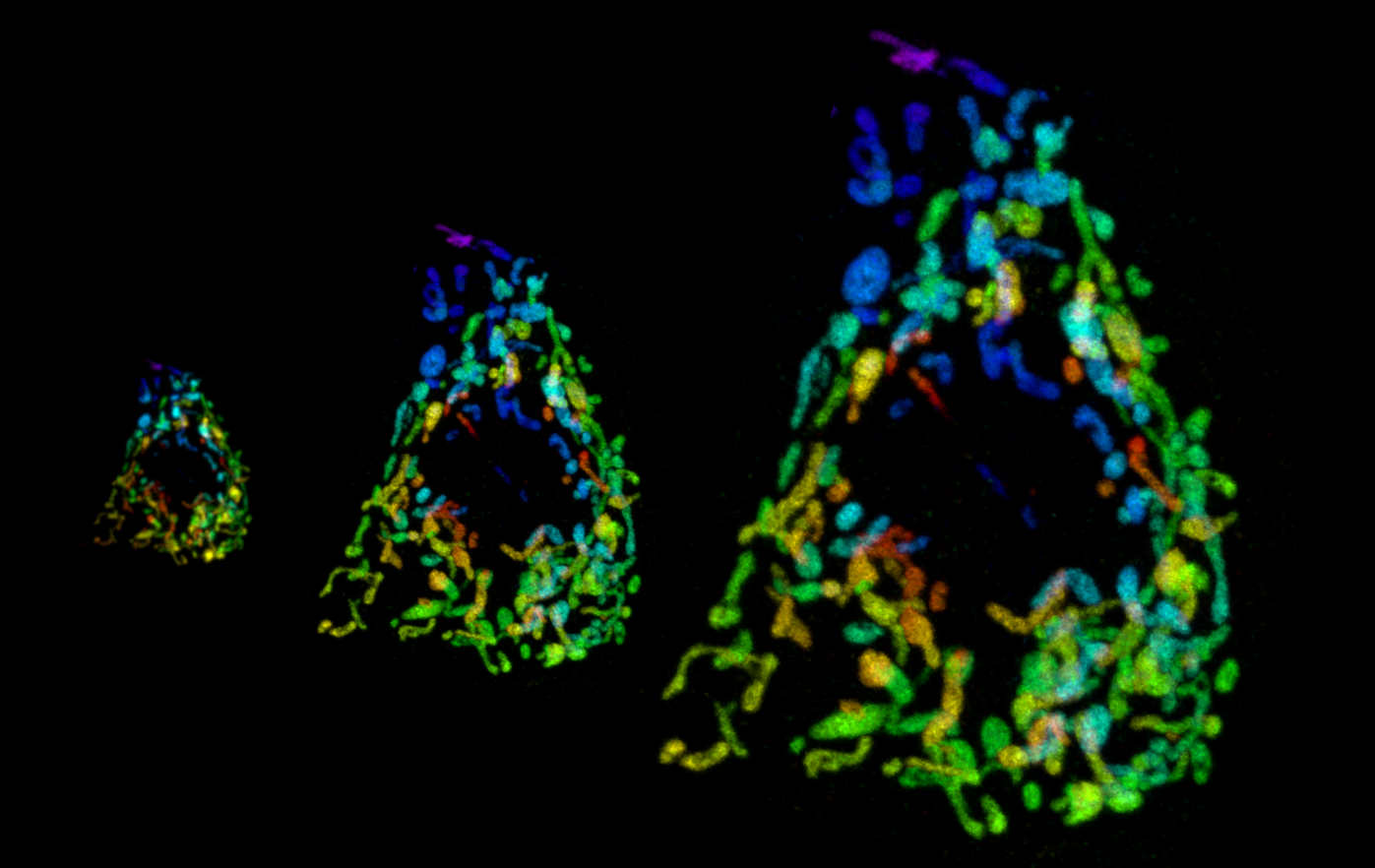Transcriptomics of sensory neurons
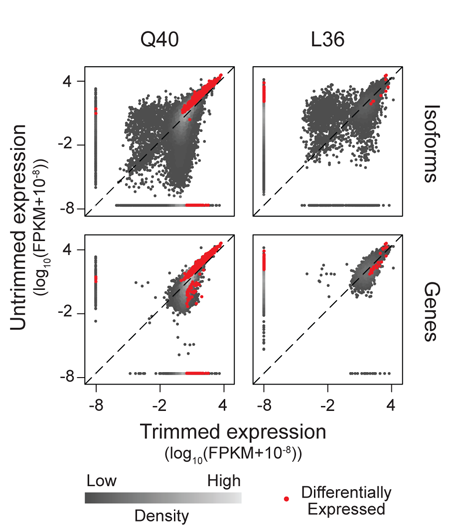
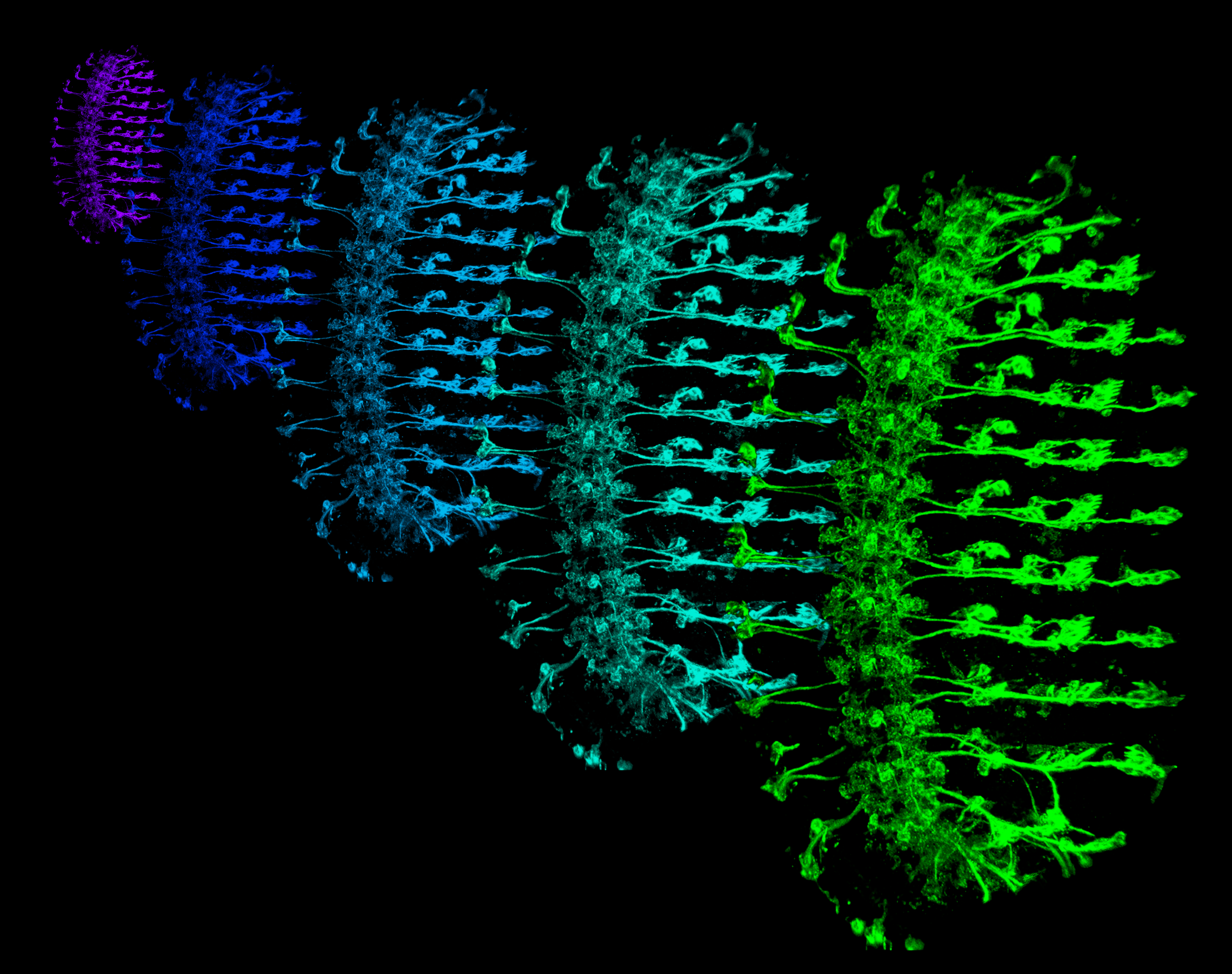
Together with Dr. Charlie Kim we have developed a robust workflow for gene expression analysis of Drosophilla neurons and non-neuronal cells that is compatible with microarray or RNAseq platforms. We have made extensive use of these approaches in our studies of growth control in sensory neurons as well as our studies of neuron-substrate interactions. Currently, we are adapting these approaches for the study of somatosensory circuitry.
Homeostatic Control of Ion Channel Expression
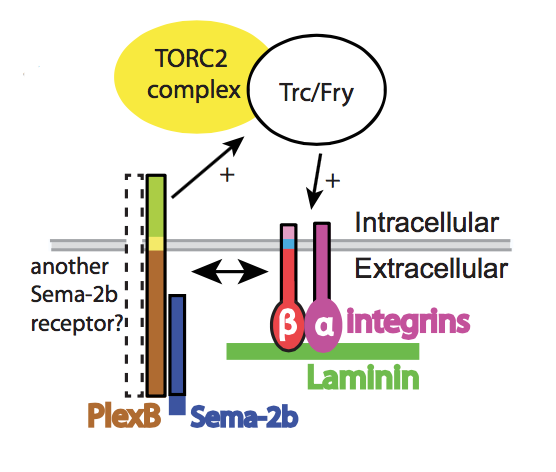
For our studies of dendrite patterning, we developed a robust pipeline for expression profiling of Drosophila neurons with single cell type resolution. Together with the labs of Grae Davis and Charlie Kim, we used these approaches to identify one of the first molecular mechanisms that participates in the homeostatic control of potassium channel gene expression. Using mutual information modeling of our gene expression data, we found that a single transcription factor (Kr) is specifically coupled to a single ion channel (Shal) and largely responsible for a gene expression program that compensates for loss of that channel. Our study therefore provides some basic principles of ion channel compensation (fidelity, specificity, selectivity, involvement of multiple channels). We hope to extend these findings by discovering additional expression relationships between ion channels.